Web-server usage
A user can choose between two options: Make predictions or Design a GPCR. The first option allows to predict the coupling preferences for input sequences, both Wild Type(WT) or mutated, as well as to visualize the sequence and structure features that are more significantly associated to a given coupling. The second one exploits instead feature information to automatically suggest the mutations more likely to switch a particular coupling, as chosen by the user. In both cases, the input can be an identifier(s) (UniProt ID/Gene names), mutations, and/or the whole sequence(s) in the form. Try out an example!
Input format and options
Precog requires a list of protein accessions (UniProt or Gene symbols), whole sequence in FASTA format, or mutations, by simply appending the positional information (e.g. D154R), separated by a forward slash (/), to the protein ID.
On the forms there are some additional options:
Mutations: Set of mutations
FASTA format: A protein specified as whole sequence
Select familes: Tick the G-protein families you had like your input to couple (for Design a GPCR option only)
Use InterPreTS: Tick this box if you had like to use InterPreTS (as a structural feature) also as a feature to predict couplings. Enter the rand value. Higher the rand, higher is the accuracy and more time it takes to run predictions
Probability cutoff for mutation difference: Enter the probability cutoff i.e. P(Mutation) - P(Wild type). Only mutations having coupling difference with at least G-protein to more than this wil shown
P(Coupling): Minimum difference that must be between the probability of variant and wild type (VAR-WT) of the input with at least one G-protein of the selected families. Value range [0.1, 0.9]. Default is set to 0.25. (for Design a GPCR option only)
P(Non-Coupling) Minimum difference that must be between the probability of wild type and variant (WT-VAR) of the input with at least one G-protein of the not-selected families. Value range [0.1, 0.9]. Default is set to 0.25. (for Design a GPCR option only)
Submit: Execute the entries
Clear: Clear the form
Upload file: Upload a file containing proteins/mutations/whole-sequences
Output page
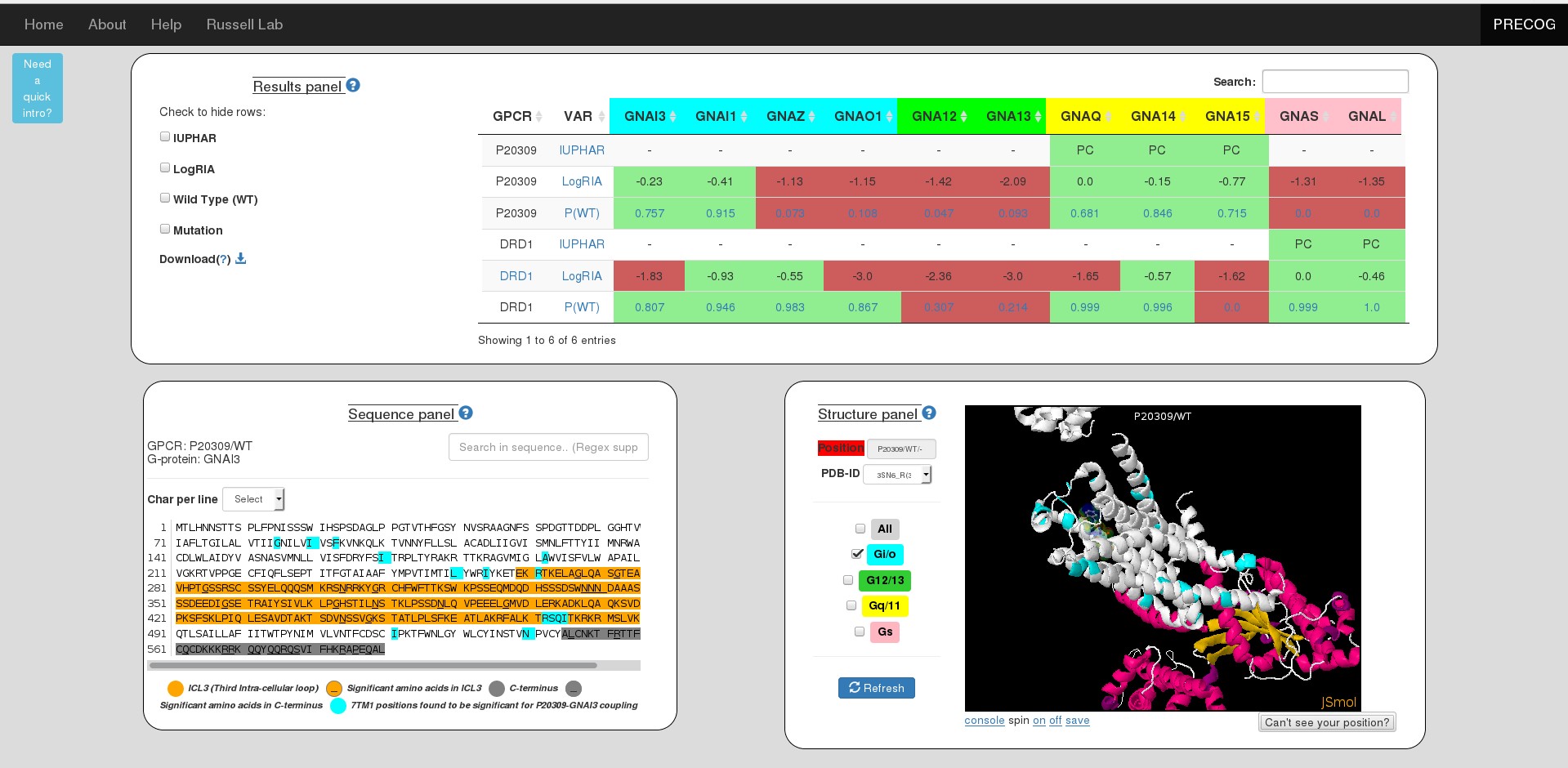
Results panel
The table here represents coupling probabilities predicted by PRECOG as well information known from IUPHAR and the Shedding Assay experiment.
Columns:
GPCR Input name(s) provided by the user.
VAR Type of variants - (a)Wild type P(WT); (b)Mutation P(Mutation); (c) IUPHAR and; (d)LogRAI
GNAI3 through GNAL Coupling probabilties predicted by PRECOG (for wild type and mutations)
Rows with:
VAR = IUPHAR display information known about the given GPCR known from IUPHAR.
VAR = LogRAI display information known about the given GPCR from the Shedding Assay experiment. The coupling values (greater than -1.0) are shown in green while the uncoupling ones are shown in red.
VAR = P(WT) display coupling probabilites of the given GPCR predicted by PRECOG.
VAR = P(Mutation) (eg: A623I - Alanine at sequence position 623 mutated to Isoleucine) display coupling probabilites of the given GPCR's mutation predicted by PRECOG. The coupling probabilites (greater than 0.5) are shown in green while the uncoupling ones (less than 0.5) are shown in red.
Click on probabilities in VAR = P(WT/Mutation) to view the GPCR in the Sequence and Structure panels.
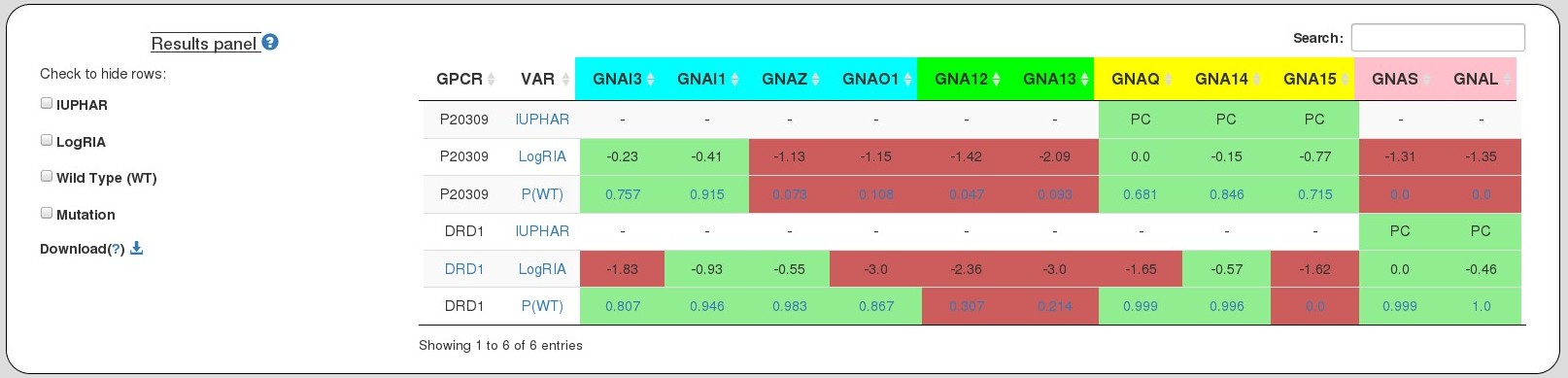
Sequence panel
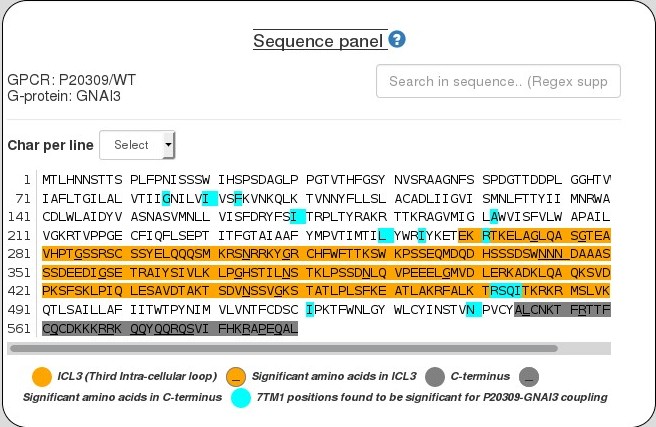
This panel shows sequence of an input GPCR, which can be selected by clicking on any of the LogRAi values or predicted coupling probabilites in the panel above (Results panel).represents coupling probabilities predicted by PRECOG as well information known from IUPHAR and the Shedding Assay experiment.
Click a significant position to view it on a structure in the Structure panel and to view its amino acid composition in a modal.
Structure panel
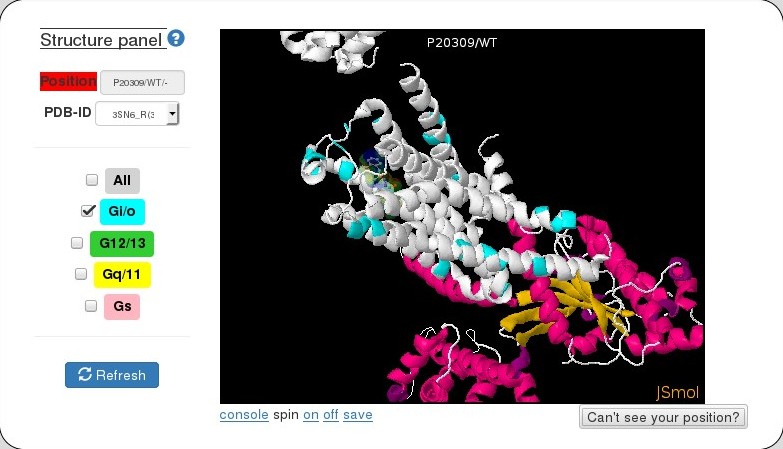
This panel shows a mutation or a significant position of given GPCR mapped onto a structure (available from PDB).
Once a mutation or significant position in selected, the PDB structure that matches best to the given GPCR (based on sequence identity using PSI-BLAST) is chosen and displayed. However, the user has option of choosing a different PDB structure from the dropdown menu (PDB-ID). The values in the paranthesis represent percentage of sequence identity of given GPCR with the PDB-ID. PDB complicies with G-protein are highlighted in coral. Contacts between 7TM positions derived using a consensus network analysis are shown as links with option of displaying the ones that are above a desired frequency.
In addition, the user also has the option of displaying significant positions of more than one G-protein families using the checkboxes and then clicking on Refresh.
Labels within the structure:
PDB Name of PDB complex displayed
Chain GPCR chain identifier of the PDB
7TM1 Corresponding 7TM domain position
BW Corresponding Ballesteros-Weinstein numbering
SEQ Selected position
Family Family for which the highlighted position is significant
Download
Download is available as TSV format. This gives information of the input and prediction probabilties by PRECOG.
Column information
Uniq_ID: A unique identifier assigned to your search by Precog
GPCR: Given entry name
VAR: Amino acid positional information of the variant (e.g. D154R)
- P(WT): predicted probabilities of the Wild type sequence of the input
- Mutations {eg: P(A263I)}: Predicted probabilites of the queried mutation (if provided)
- IUPHAR: Information known from IUPHAR
- LogRAi: Values known from the shedding assay. (Non-coupling GPCRs < -1.0 < Coupling GPCRs)
[GNAI3 - GNAL]: Probabilities (in case of Wild type and mutations) or known information (in case of IUPHAR and LogRAi) of the entry for the corresponding G-protein
7TM1_POS/BW/ALN_POS: Values between '/' (Only applicable in case of mutations)
- 1st: The 7TM1-domain position of the mutation
- 2nd: Ballesteros-Weinstein numbering of the position affected
- 3rd: Internal numbering used by Precog
- [4th:]: The G-proteins(if any) affected by the mutation
Mutation_Info: Disease related information(if any) of the mutation retrieved from Uniprot
Browser compatibility
| OS | Version | Chrome | Firefox | Safari |
|---|---|---|---|---|
| Linux | Ubuntu 16.04 | not tested | Quantum 64.0 | n/a |
| Linux | CentOS 7.2.1511 | not tested | ESR 52.6.0 | n/a |
| Windows | 10 | v71 | not tested | n/a |
| MacOS | Mojave 10.14.2 | v71 | v64.0 | 12.0 |
Contact
Rusell Lab at the University of Heidelberg, Germany
Gurdeep Singh: gurdeep.singh@bioquant.uni-heidelberg.de
Francesco Raimondi: francesco.raimondi@bioquant.uni-heidelberg.de
Rob Russell: robert.russell@bioquant.uni-heidelberg.de
Inoue Lab at Tohoku University, Japan
Asuka Inoue: iaska@m.tohoku.ac.jp
